| Research
|
Jerry Tuskan says that his lifelong appreciation for trees began in the upper peninsula of Michigan and has been a constant influence, taking him down a path to a very exciting research career focused on the genetics of tree growth and development. Tuskan came to DOE's Oak Ridge National Laboratory in 1990 to work on an early biomass feedstock program investigating alternative fuels. Coincidentally, the Human Genome Initiative happened to wrap up at about the same time as his program was ending and Tuskan saw an opportunity in the slowing sequencing facilities that had been devoted to sequencing the human genome. He submitted a proposal to sequence the first tree genome, and is now using that sequence to improve biomass production and carbon sequestration. Accomplishing the intricate job of sequencing the massive Populus genome, which has 1.5 times more genes than the human genome, required the concerted efforts of more than 80 collaborators. "It is the most complex genome sequenced in the world. The Populus genome has undergone a duplication event, so assembling the sequence was like putting together a jigsaw puzzle of a tree reflected in a lake; you have to figure out whether the piece of the sequence belongs in the reflection or in the original,” says Tuskan. A plant can process carbon in several different ways, incorporating it into cell walls or using it for energy production and releasing it back into the environment. Identifying the genes governing these pathways can help researchers maximize the amount of carbon that is stored stably and for long periods of time in the form of cell walls, making increasingly energy-dense biomass and removing carbon from the atmosphere. According to Tuskan, these are heady times for studying tree growth and development: “The truth is, we really don’t know what makes a tree a tree, but we’re getting closer and closer.”—Sarah WrightSubmitted by DOE's Oak Ridge National Laboratory |
|||||||||||||||||||||||||||
|
Check out the joint Fermilab/SLAC publication symmetry.
|
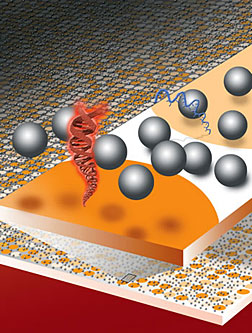
Easy to read gene chips could revolutionize medical diagnosticsThe dream of personalized medicine—in which diagnostics and treatment decisions are based on a patient’s genetic profile— may soon be expanded beyond the wealthiest of nations with state-of-the-art clinics. Berkeley Lab researchers have invented a technique in which DNA or RNA microarrays, often referred to as “gene chips,” can be read and evaluated without the need of elaborate chemical labeling or sophisticated instrumentation. Based on electrostatic repulsion—objects with the same electrical charge repel one another—the technique is relatively simple and inexpensive to implement, and can be carried out in a matter of minutes.
“One of the most amazing things about our electrostatic detection method is that it requires nothing more than the naked eye to read out results that currently require chemical labeling and confocal laser scanners,” said Jay Groves, a chemist with Berkeley Lab’s Physical Biosciences Division, who led this research. “We believe this technique could revolutionize the use of DNA microarrays for both research and diagnostics.” Gene chips have become one of the most powerful tools for gene-expression profiling and the identification of mutations, and for diagnostics in patients afflicted either by multiple diseases or drug-resistant strains of diseases. Until now, however, gene chip use has been limited because current techniques for reading them depend upon fluorescence detection, a demanding methodology that requires time-consuming chemical labeling, high-power excitation sources and sophisticated instrumentation for scanning. Groves and his colleagues have developed a technique in which a fluid of electrically-charged microscopic glass beads is dispersed across the surface of a DNA microarray. Observing the Brownian motion of the beads provides measurements of the electrical charges of the DNA molecules that can be used to interrogate millions of DNA sequences at a time. What’s more, these measurements can be observed and recorded with a simple hand-held imaging device—even a cell phone camera will do.Submitted by DOE's Lawrence Berkeley |