- Number 402 |
- December 2, 2013
Lost in translation: Gene expression changes don’t always alter protein levels
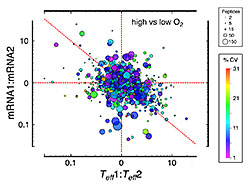
The relative contribution of transcription
(vertical axis) versus translational efficiency
(horizontal axis) to changes in protein
expression. The change in both directions
shows that transcription is not the sole
determinant of altered protein levels.
To determine if the common assumption – changes in specific messenger RNA (mRNA) levels are always accompanied by commensurate changes in the encoded proteins – is true, scientists at DOE's Pacific Northwest National Laboratory examined Shewanella oneidensis MR-1 grown under steady state conditions at either 20% or 8.5% oxygen. Using a combination of quantitative proteomics and a next-generation sequencing technology, they generated high-confidence data on more than 1000 mRNA and protein pairs. Surprisingly, they found that changes in the expression of proteins in response to altered oxygen levels were caused primarily by differences in the translational efficiency of the mRNAs rather than changes in the mRNA levels.
For example, when oxygen levels were lowered, 28% of the detected proteins showed at least a twofold change in expression. Altered transcription levels appeared responsible for 26% of the protein changes, altered translational efficiency appeared responsible for 46%, and a combination of both were responsible for the remaining 28%.
Interestingly, changes in translational efficiency were significantly correlated with the pattern of codons in genes, which encode different amino acids. Because measurable transfer RNA pools, which read the codons, also changed in response to altered oxygen levels, this suggests that changes in the translational efficiency of genes are caused, in part, by alterations in the abundance of molecules that decode the mRNAs.
“To understand how microbes work together in communities, it’s important to understand the functional state of their member species,” said Dr. Steven Wiley, PNNL Laboratory Fellow, who led the work. “Metagenomics analyses have provided important data on the functional potential of each community species, but their actual functions are highly dependent on their pattern of protein expression, which, in turn, is modulated by the local microenvironment of each cell.”
How cells change the expression of proteins in response to their environment is one of biology’s most fundamental questions. Theoretically, changes in the synthesis of specific mRNAs can give rise to altered protein levels, but the multitude of steps between transcription and translation provide many different regulatory opportunities. Advances in high-throughput sequencing technologies and mass spectrometry now allow scientists to look globally at the multiple steps that control protein expression levels. These approaches provide the potential for understanding larger, systems-level regulatory mechanisms that can provide insight into fundamental biological design principles. The integrated and quantitative approach developed here to identify the level at which proteins are regulated provides a foundation for more detailed mechanistic studies to understand how cells control protein expression and for explorations of why translational efficiency changes under different cell environments.
This research was supported by DOE’s Office of Biological and Environmental Research Genomic Science Program, under the PNNL Foundational Scientific Focus Area. The research was performed in http://www.emsl.pnnl.gov/emslweb/EMSL, a BER national scientific user facility. This work was also funded in part by the National Institutes of Health.[Kristin Manke, 509.372.6011,
kristin.manke@pnnl.gov]
