- Number 342 |
- July 25, 2011
Model helps pinpoint bacterial genes that capture the sun’s energy
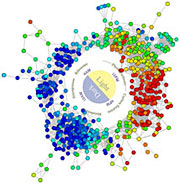
Researchers developed this
wreath-like graph to visualize
all the genes being expressed
by Cyanothece in 24 hours. The
graphic revealed the complex
genetic network that enables
Cyanothece to switch its cell
between photosynthesis and
nitrogen fixation as the day
turns to night.
In the battle to reduce our need for fossil fuels, scientists at DOE’s Pacific Northwest National Laboratory, Washington University in St. Louis and Purdue University are turning to an unassuming ally: known as Cyanothece 51142. This organism is commonly referred to as a cynaobacteria. The researchers developed a computer model that can predict which of the organism's genes are central to capturing energy from sunlight and using it to produce fuel.
"Our model is the first of its kind for cyanobacteria," said Jason McDermott, a PNNL computational biologist and the study’s lead author. "Previous models have only zoomed in on specific aspects of cyanobacteria. Ours looks at the entire organism to find out what makes Cyanothece tick."
Cyanothece makes sugars, which could be the basis for alternative fuels, when there's daylight. Then, it spends the night breaking down that sugar to fix nitrogen and to produce other compounds. The Cyanothece is unusual because the same cell switches between these functions every 12 hours.
Submitted by DOE's Pacific Northwest National Laboratory
