Archive Site Provided for Historical Purposes

Sponsored by the U.S. Department of Energy Human Genome Program
Human Genome News, September-December 1995; 7(3-4):13
Version 6.0 Release Expected in January 1996
When the Genome Data Base began public operation at Johns Hopkins University in 1990, researchers had been mapping the human genome for several decades. In keeping with the Human Genome Project's mission to make mapping data freely accessible to the scientific community, GDB set about curating the existing data and making the information available to those engaged in the mapping effort. The first release, GDB 1.0, debuted in September 1990 at HGM 10.5, offering a character-based interface with pull-down menus. It provided editorial functions, text-based genetic maps, a means for managing literature references, and data on probes and polymorphisms.
Since its inception as the official repository for mapping data in support of the genome project, GDB's efforts always have been dedicated to improving access and creating a data model that would accommodate different mapping methodologies and the data they generate. Allowing the user to view the data in a more meaningful way one that reflected the richness of the biological data became an important focus for GDB. This resulted in a series of releases aimed at making the task easier for the user to view and submit data. A statement in the report of the informatics committee at the August 1991 HGM 11 (when GDB 4.1 was in use) reads, "Graphical tools to display, analyze, and compare this information will be developed in the future." The future has finally arrived.
GDB has been aware of the need to provide a graphical representation of stored maps. An associated need is the logical imperative to provide an easier way for the user to edit stored maps and data and create new maps. With these two goals in mind, developers set out to create a set of applications that allow users to create, view, and edit maps off line, submit data directly, and retrieve data using more-flexible query tools.
GDB Version 6
The GDB 6 release will represent a fundamental change in both database structure and user interface. Unlike past major GDB releases that were issued infrequently and included many changes, the new series will be released incrementally to shorten the time between releases and add features as they become available. GDB 6.0 will take advantage of the user's familiarity with WWW to introduce a graphical interface. Subsequent releases will add additional query and editing functions.
A generic Web browser based on Stan Letovsky's Genera technology will allow users to query the database as before. Furthermore, this enhanced interface will enable customized map queries based on order and distance; this was not possible previously. Entering data by hand, including map data, will be accomplished through the Web interface. Once entered, data can be edited and displayed in a graphical format through the GDB map viewer.
Later GDB Releases
Later releases in the GDB 6 series will allow users to create and edit maps through the map editor, which has evolved from the map-viewer component of an applications suite. All users will be able to submit data directly, edit their own previous submissions, and annotate (but not change) the submissions of others. The ability to prepare submissions off line reduces dependence on the Internet, a plus as net traffic steadily increases. Map-viewer software will provide an export function allowing map customization with graphical drawing programs.
Maps that currently reside in GDB will be viewable through the map viewer. Cytogenetic, linkage, content-contig, and radiation-hybrid maps will be supported initially; other types will be added later. Consensus and observed maps will be available, providing both an authoritative source for the novice as well as the most-current mapping data to produce an integrated map for the experienced user. Clicking on an object in the map will display automatically the details of that object in the Netscape Web browser.
Because of the enhanced editorial features of GDB 6, the ability to identify data modifications or altered links to other data objects becomes an extremely important feature. Users will be able to specify and view map versions and display an edit trail of specified versions.
Behind the scenes, the database architecture has been significantly altered, moving from a monolithic to a modular structure. Previously, one large database of objects was stored in tables and linked in various relationships; now, data are stored in several smaller databases. The "object broker" at the core facilitates relationships among objects contained within the modular databases. This model allows other modules to be added without altering the underlying schema.
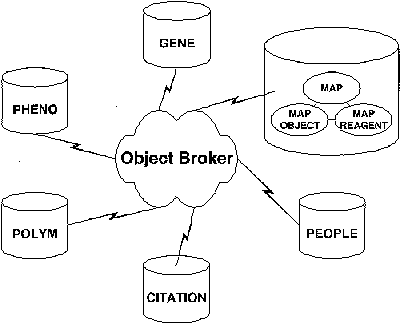
Fig. 1. GDB modular database architecture allows modules to be added without altering the underlying schema. The object broker provides a uniform interface across multiple databases.
Database Federation
The use of an object broker opens up opportunities for a possible federation of genomic databases, and the new architecture allows a client to access information contained in multiple databases.
Figure 2 (below) depicts various client-server relationships as they will function conceptually in GDB 6.0 and a database federation.
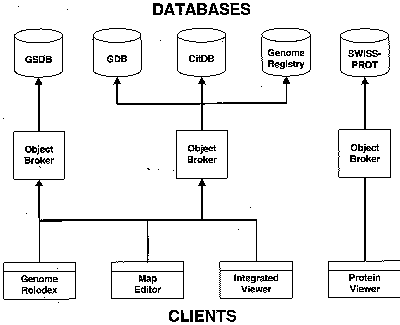
Fig. 2. Client-server relationships in GDB 6.0 and in a database federation.
GDB began assigning accession numbers to all database objects in July 1993. These external identifiers were a first step in linking objects in one database with relevant genomic data stored in other databases, regardless of format or location. The object broker will provide a means of retrieving linked data from multiple databases participating in the federation. The current version of GDB includes links through its Web server to such other databases as the Genome Sequence Data Base, Mouse Genome Database, and SWISS-PROT. This capability will be expanded in GDB 6.
Mapping data continues to increase as the genome project progresses. The release of GDB 6 will provide a new and improved method of navigating and viewing data and relating it to other new genomic insights.
Return to the Table of Contents
The electronic form of the newsletter may be cited in the following style:
Human Genome Program, U.S. Department of Energy, Human Genome News (v7n3).
The Human Genome Project (HGP) was an international 13-year effort, 1990 to 2003. Primary goals were to discover the complete set of human genes and make them accessible for further biological study, and determine the complete sequence of DNA bases in the human genome. See Timeline for more HGP history.
Published from 1989 until 2002, this newsletter facilitated HGP communication, helped prevent duplication of research effort, and informed persons interested in genome research.